The summary table used to generate the plot of repeat content against genome size is available for download alongside individual RepeatMasker summary tables for each assembly on download.lepbase.org.
Repeat content
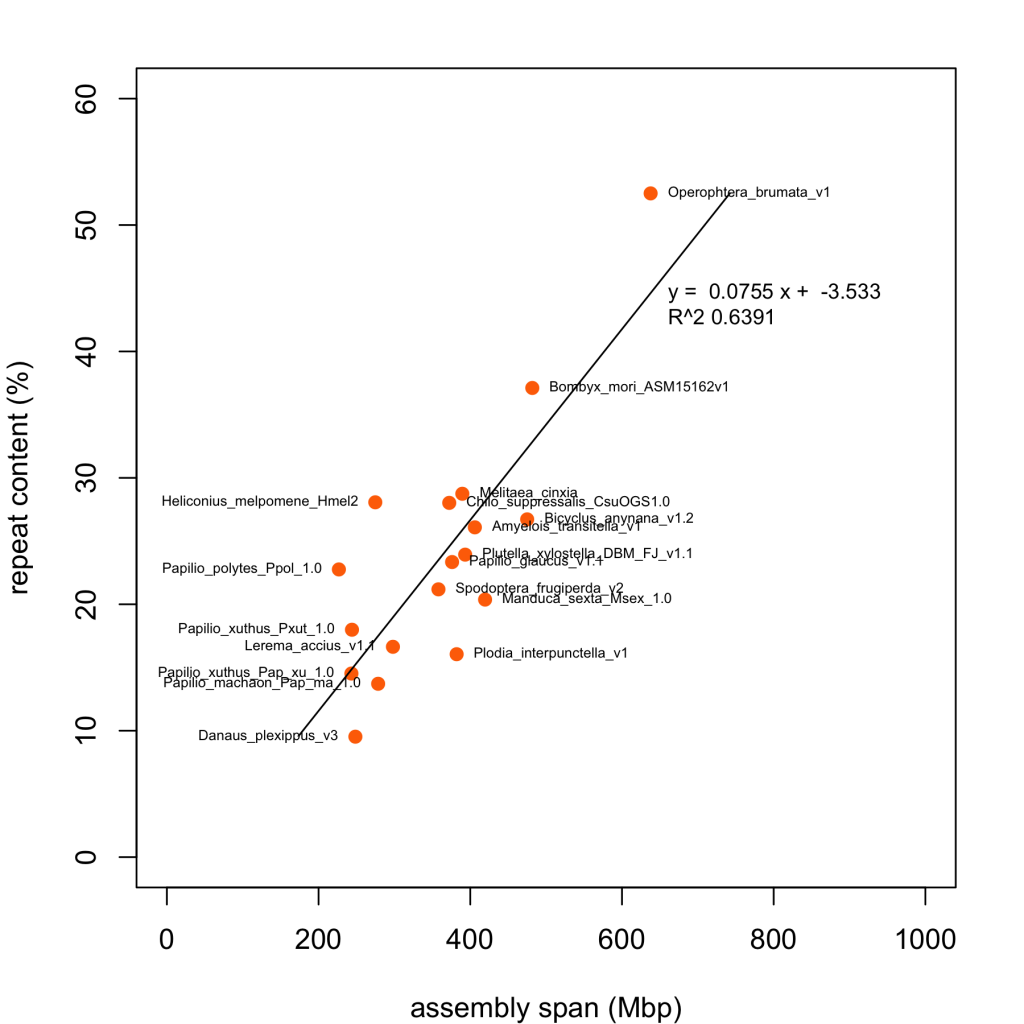
Inspired by the recent review by Canapa et al. , here’s a plot showing the relationship between repeat content and genome size for most of the Lepidoptera genome assemblies in Lepbase. Repeat content for each assembly was calculated with the same standardised pipeline using RepeatModeler /RepeatMasker , described at blaxter-lab-documentation.readthedocs.org.
This confirms Operophtera brumata v1 as having the highest repeat content of any publishe Lepidopteran genome and Danaus plexippus v3 the lowest. Assemblies plotted well below the regression line may have lower than expected repeat content due to genuine low genomic repeat content or collapsed repeats in the assembly.
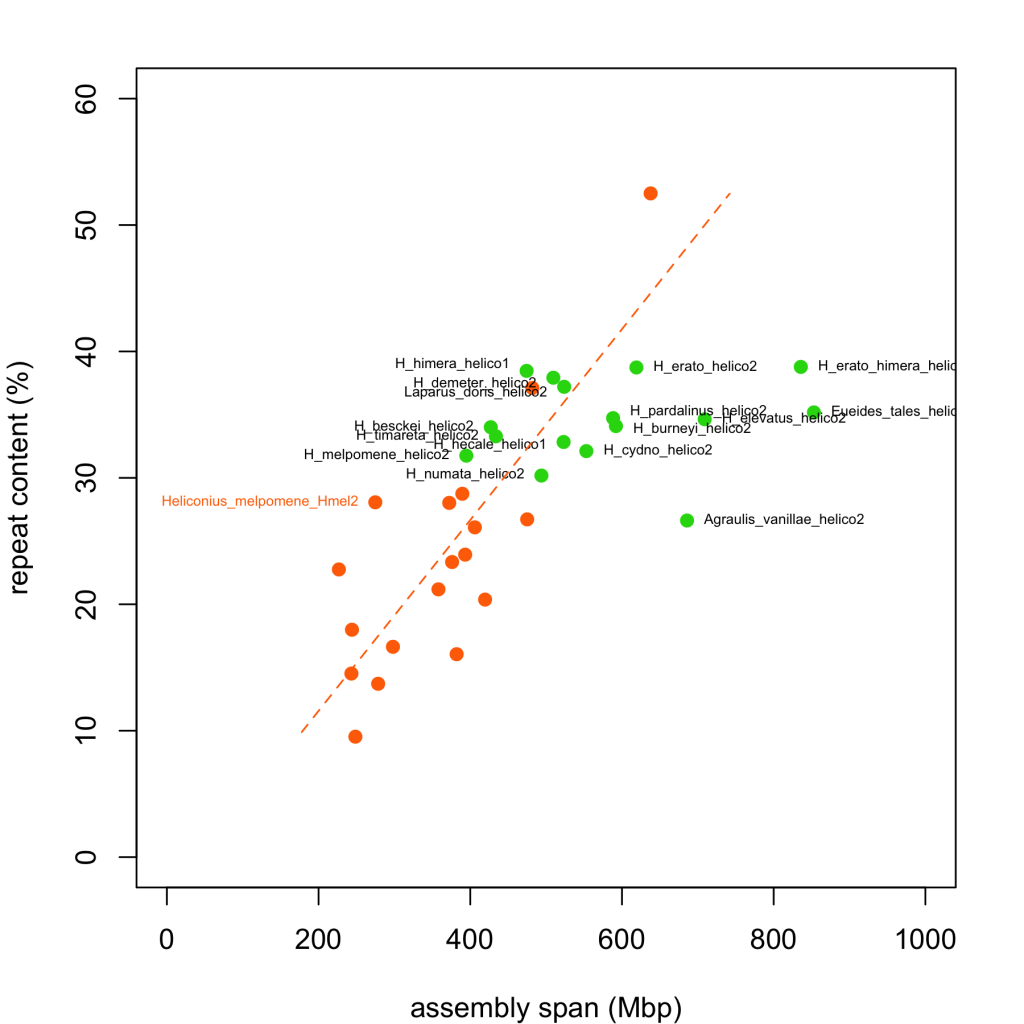
For completeness the plot below shows the relationship between assembled genome size and annotated repeat content for the highly fragmented Heliconiine DISCOVAR assemblies, masked using the Hmel2 repeat library. This should be interpreted with caution as there are likely phylogenetic artefacts of using a single species repeat library in addition to any assembly artefacts.