Version 4 of Lepbase, the hub for Lepidopteran genomes, not only includes several new genome assemblies and annotations, but also showcases a re-engineered infrastructure that will enable other groups to easily set up their own Ensembl-based genome hubs in the future.
New datasets
New genomes with gene models:
Plutella xylostella pacbiov1 – Alistair Darby’s Lab
Heliconius erato lativitta v1 – Robert Reed’s Lab
Additional assemblies or annotations for existing species:
Previously, Lepbase v3 had hosted NCBI RefSeq annotations for Papilio xuthus and Papilio polytes. RefSeq annotations are independent gene predictions generated by the NCBI, but may include sequence data that are not present in the genome assemblies because they use additional information from independent transcripts. In Lepbase v4 we have also included the original gene predictions from the Fujiwara lab that sequenced these genomes:
Papilio xuthus Pxut 1.0
Papilio xuthus Pxut 1.0 Refseq
Papilio polytes Ppol 1.0
Papilio polytes Ppol 1.0 Refseq
The Heliconius melpomene genome assembly version remains the same (Hmel2), but the annotation has now been updated to include Braker 1.0 predictions based on several independently sequenced RNAseq libraries, in collaboration with Chris Jiggins’ lab. Manual annotations and old gene names have been retained where the overlap is unambiguous.
Heliconius melpomene melpomene Hmel2
Assembly-only species:
Several new assembly-only species were also added to Lepbase v4. Although no gene prediction sets are available for these species, you can download the genome fasta files at download.lepbase.org/v4/sequence and do BLAST searches against them at blast.lepbase.org.
Low-coverage draft genome assemblies from Ferguson et al (2014) :
Callimorpha dominula
Cameraria ohridella
Hepialus sylvina
Pararge aegeria
Polygonia c-album
Glyphotaelius pellucidus (caddisfly outgroup)
The Heliconius genome consortium has released 24 new Heliconiine genome assemblies using the w2-wrap contigger and a-scaff tools, courtesy Jim Mallet and Bernardo Clavijo’s group at Earlham Institute. Some of the samples are species crosses or were contaminated, but we decided to host them as separate genome assemblies to make them easier to search against:
Agraulis vanillae
Dryas iulia
Eueides tales
Heliconius besckei
Heliconius burneyi
Heliconius cydno
Heliconius demeter
Heliconius elevatus
Heliconius erato mother
Heliconius erato x himera f1
Heliconius hecale
Heliconius hecale old
Heliconius hecalesia
Heliconius himera
Heliconius himera father
Heliconius melpomene
Heliconius numata
Heliconius pardalinus
Heliconius sara
Heliconius telesiphe
Heliconius telesiphe contaminated
Heliconius timareta
Laparus doris
Neruda aoede contaminated
We are also planning to make these assemblies more useful by running a comparative gene prediction across all the heliconiines. The first step towards this is to compute a whole-genome alignment using Progressive Cactus, which we have made available for download at download.lepbase.org/v4/progressivecactus
New and updated analyses
Gene trees
We generated a comprehensive gene orthology analyses spanning 23 genomes (21 species, but with two sets of genomes/gene-predictions for Heliconius erato and Plutella xylostella). Rather than using the Ensembl Compara pipeline as is, we used current best practices in gene-tree reconstruction such as:
- OrthoFinder instead of OrthoMCL for homologue clustering
- MAFFT for protein sequence alignment
- Noisy for automated filtering of multiple sequence alignments
- RAxML for phylogenetic inference
- NOTUNG for gene-tree/species-tree reconciliation.
Over 12,000 OrthoFinder clusters were processed into Gene Trees which can be accessed from their constituent gene pages in the Lepbase Ensembl browser.
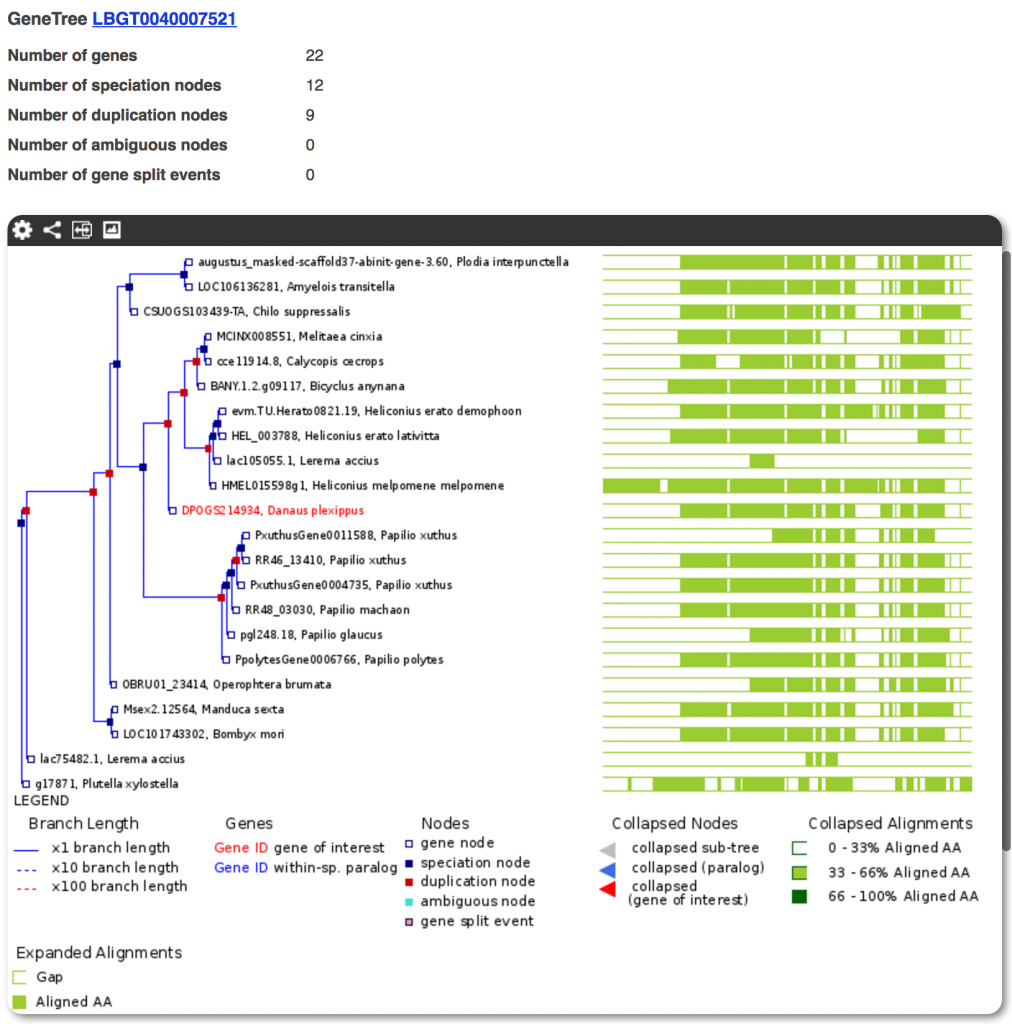
The goal of this analysis was to provide a context for each gene. However, as with all automated analyses, it may contain artifacts due to differences in gene prediction methods or due to generic parameters used in the pipeline. If you are interested in a highly accurate reconstruction of a specific gene family, you can download all the sequence and alignment data for a given gene and its homologues, and redo the tree using your preferred phylogenetic pipeline. The specific steps and evaluations for each part of our gene-tree pipeline will be described in forthcoming technical notes.
RepeatMasking
One of the key features of Lepbase is that we provide consistent analyses across all genomes using the same software and database versions and parameters. All genome sequences were masked using RepeatMasker 4.0.6 with the built-in arthropod repeat database. Previous Lepbase releases used RepeatModeler to generate species-specific repeat libraries. However, we found that RepeatModeler risks masking recently expanded gene families, therefore we took a more conservative approach and did not use RepeatModeler for this v4 release.
download.lepbase.org/v4/repeatmasker
Functional annotation using blastp and interproscan
We also provide functional annotations for protein-coding genes in all species with gene models using BLASTP against the SwissProt database, and using all the tools provided by InterProScan.
download.lepbase.org/v4/blastp
download.lepbase.org/v4/interproscan
New infrastructure and features
Lepbase.org previously ran on Linux virtual machines because of the complex dependencies of the various software programs involved. For Lepbase release v4, we have successfully migrated all the services, including the import and annotation pipelines for new genomes, to a Docker-based container infrastructure.
None of this affects how the service looks to the outside world, but it makes it much easier for us to upgrade the software as new versions of Ensembl and other software are released. We will also be able to easily scale up individual services to meet the growing number of users.
Although all our code is public already (see github.org/lepbase and easy-import.readme.io), we plan to document it extensively in the coming weeks, so that other groups can rapidly and easily set up their own taxon-specific Ensembl-based genome hubs using our Docker infrastructure.

