Lepbase release 3
We are pleased to announce the third release of our Lepidopteran genome browser including new datasets, updated analyses, a new look, and some code changes that will help us to add new data between major releases in the future.
New datasets
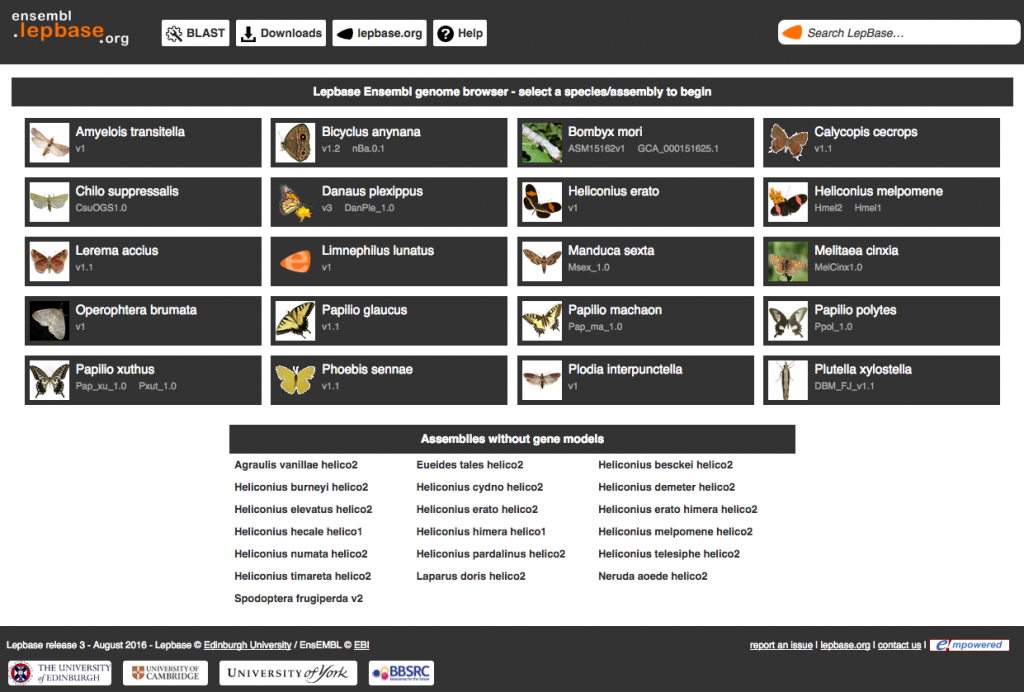
We’ve added assemblies for 4 species, bringing our tally up to 43 assemblies in 36 species, all available on our Ensembl genome browser (ensembl.lepbase.org), BLAST interface (blast.lepbase.org) and downloads server (download.lepbase.org). New datasets include:
-
 Heliconius erato: the red postman butterfly
Heliconius erato: the red postman butterfly  Calycopis cecrops: the red-banded hairstreak butterfly
Calycopis cecrops: the red-banded hairstreak butterfly Phoebis sennae: the cloudless sulphur butterfly
Phoebis sennae: the cloudless sulphur butterfly Limnephilus lunatus: the caddis fly, as a representative of the sister group to the Lepidoptera
Limnephilus lunatus: the caddis fly, as a representative of the sister group to the Lepidoptera Bicyclus anynana v1.2 preliminary gene models
Bicyclus anynana v1.2 preliminary gene models
Updated analyses
We run a standard (and consistent) set of analyses across all assemblies/gene sets, including InterProScan, Blastp against SwissProt, RepeatModeler/RepeatMasker. Blastp and InterProScan allow for text searches of gene models and proteins based on domain names or known genes in other species. These analyses are also available as bulk downloads: Blastp and InterProScan.
We have also updated our comparative pipeline to produce more accurate orthology predictions across the gene sets that were available in our previous release. These are all available at v2.ensembl.lepbase.org (e.g., gene tree for Bombyxin showing all orthologs and paralogs). It’s taken a while to work out the import process for adding these to the Ensembl Compara database so we decided to release the genomes that we have for now and we’ll update the gene trees to include the recently added v3 assemblies soon.
Whole genome alignments are also on the way – for now the multi-species alignments we have run are available at download.lepbase.org/current/wga/
New look
We’ve tried to make our main portals a little more consistent.
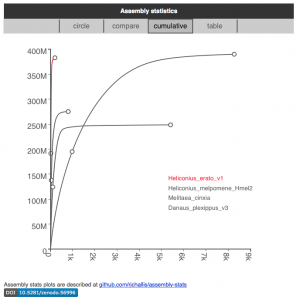
We’ve also added more interactive data visualisations to the species pages on ensembl.lepbase.org. We now base these on a standardised file structure so we can offer alternative views, such as table and cumulative frequency distribution views for the assembly statistics. The code for these new visualisations is also available (see the link below each image).
Code changes
We don’t expect the average Lepbase user to be particularly interested in the code we use but we’re quite excited about the changes we’ve made to our Ensembl import scripts to create an easy-import pipeline that makes it very simple to set up and add species, not just to Lepbase but to any custom Ensembl instance. We expect this to make it simple for us to add new data as they become available so if you are working on a project that you would like to see included then please get in touch. We’re already starting to use Ensembls more widely in the Blaxter Lab and we hope our code will be useful to anyone who has been thinking about setting up an Ensembl-based website – check out the documentation at easy-import.readme.io to see just how simple setting up your own Lepbase could be.