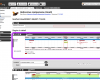
Ensembl views
After identifying a region with the Text or BLAST searches, the genome browser splits the available information into three distinct views, gene, transcript and location. Each of these views provides access to different features and annotations.
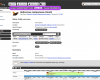

Gene trees
Updated gene trees will be available shortly.
Orthogroups and gene trees are only available on the "Gene" pages, either by clicking on the buttons in the main view or the links on the left-hand side.
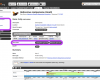

Gene tree viewer
Updated gene trees will be available shortly.
The gene tree viewer provides an interactive phylogeny making it straightforward to access orthologues and paralogues of the target gene by clicking the labels on the leaf nodes.
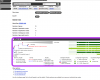

BLAST genes
The sequence for a gene can be sent directly to the Lepbase BLAST server for homology searches in any of the Lepbase sequence datasets by clicking the button.
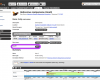

BLAST transcripts
Because of the way that information is divided between the views, transcript pages offer some of the same information as the gene pages and additional transcript-level information, such as the ability to send CDS or protein sequences to the BLAST server are also available.
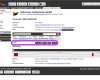

Sequences
Sequence data can be accessed directly by clicking the links on the left-hand side. Additional links to external references and alternate views are also listed in this menu when available.
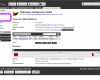

Protein displays
Proteins are stored as transcript translations so protein-level data and annotations are also accessed from within the transcript view.
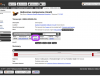

Location view
The location view provides an overview of the features annotated on a scaffold or scaffold segment. Click on the genes here to access detailed information for neighbouring genes.